Disease resilience indicators from individual feed intake data in a high-health swine nucleus
Chad Bierman, PhD, Geneticist, Genesus Inc.
Profitability for swine producers has traditionally focused on lowering cost, improving efficiency, or increasing value. An important component of efficiency is maximizing marketable product. Delivering more product to market makes economic sense, particularly when almost all input costs have already been expended. Therefore not unexpectedly, mortality and morbidity was shown to comprise the three highest metrics impacting profitability in swine production, as highlighted by Boyd (2012), because both effects contribute to less pounds of product sold (selling lighter or fewer) and significant costs in the case of mortality of pigs in the later phases of finishing. Acquiring measurements of mortality, morbidity, or associated indicator traits, allows for genetic improvement in this area, leading to increased profitability.
Feed intake (FI) traits have received much focus in swine breeding programs. Appetite drives growth rate, which increases end weight, and is also linked to robustness (Ocepek et al. 2019). Stressors such as disease are known to negatively impact FI and subsequent growth (Nguyen-Ba et al. 2020). The variability in feed intake under a disease challenge has been shown to affect performance and survival and has resulted in the confirmation of these traits as heritable and favorably genetically correlated with production and mortality outcomes under such stress (Cheng et al. 2020, Putz et al. 2019). This suggests that FI resilience traits are plausible indicator traits of disease resilience.
Feed intake traits are commonly recorded in genetic nucleus herds where populations are under direct selection. These herds most often operate under a high-health scenario to maximize the pigs’ ability to express their full genetic potential, maximize accuracy of genetic parameter estimation and genetic improvement as a result. Feed intake resilience phenotypes are therefore readily calculable, but are they heritable under this high-health situation?
FI resilience traits were computed from 3.5 million daily FI records (7,498 animals) captured between 2016-2021within a Genesus Duroc nucleus unit per methodologies recorded previously (Cheng et al. 2020, Putz et al. 2019). One of these measurements involves calculating the amount of variation for daily feed intake (kg/day) and daily feed intake duration (the amount of time spent eating) (Figure 1). Phenotypes were grouped into categories of higher or lower stress based on the average monthly temperature at the time when animals were taken off test. This partitioning provided a seasonality effect designed to separate phenotypes into stress categories based on climatic conditions. Although barn temperature and ventilation are electronically controlled, humidity and hot/cold microenvironments are difficult to overcome during climatic extremes, resulting in differing stress levels. Variance components were estimated to determine heritability and genetic relationships with measures of average daily FI (ADFI).
Figure 1. Examples of two pigs with visibly lower (A) or higher (B) variation in daily feed intake (VARFI), accompanied by their plot of variation in feed intake duration (VARDur). The amount of shaded area above and below the trendline signifies the amount of variation. Slope of the trendline is independent from the variation measured for VARFI or VARDur. These two pigs do not differ in their VARDur phenotype.
A B
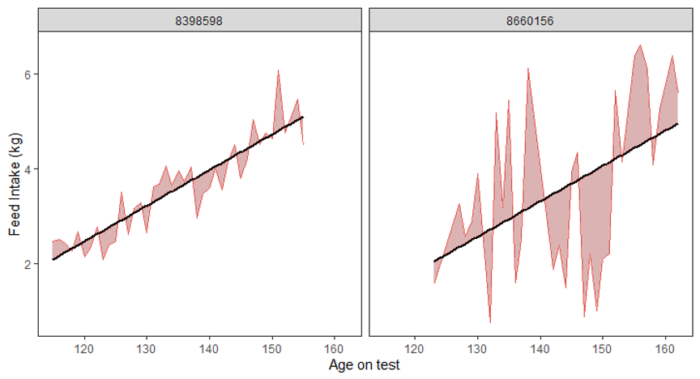
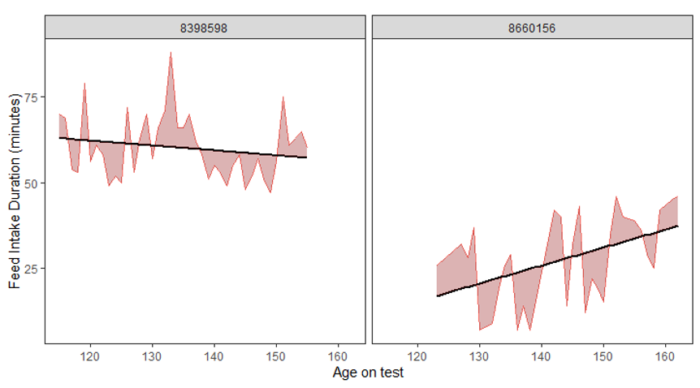
Heritability estimates were moderate, with ADFI larger for the higher stress compared to lower stress category, and the resilience traits similar between the two stress categories (Table 1). Estimates of genetic correlation (Table 2) between ADFI and the amount of variation in feed intake duration (VARdur) matched previous reports, where high variation in duration at the feeder resulted in lower ADFI. This relationship appears stronger under higher stress than under lower stress. Correlation estimates for variation in FI (VARFI) show mixed signals between stress environments, rendering the VARFI phenotype inconclusive as to its use in a breeding program. For future consideration, the standard error (SE) implies that these correlations could benefit from improved estimates, which can be achieved by accumulating more records for these FI resilience traits.
Table 1. Estimates of heritability and standard errors for performance and feed intake resilience traits under different stress categories.
| Higher stress | Lower stress | |||
| Heritability | SE | Heritability | SE | |
| ADFI | 0.46 | 0.04 | 0.35 | 0.05 |
| VARFI | 0.33 | 0.04 | 0.31 | 0.04 |
| VARdur | 0.50 | 0.04 | 0.50 | 0.04 |
VARFI = Variation in daily feed intake; VARdur = Variation in daily feed intake duration
ADFI = Average Daily Feed Intake; SE= Standard Error
Table 2. Estimates of genetic correlations (SE) between feed intake resilience traits and ADFI under different stress categories.
| ADFI | ||
| Higher stress | Lower stress | |
| VARFI | 0.00 (0.15) | 0.49 (0.16) |
| VARdur | -0.22 (0.11) | -0.12 (0.16) |
VARFI = Variation in daily feed intake; VARdur = Variation in daily feed intake duration
ADFI = Average Daily Feed Intake; Standard Errors (SE) in parentheses
In conclusion, FI resilience traits are heritable in a high-health nucleus setting. This is important because it provides an opportunity for Genesus to create novel phenotypes from existing data to potentially add to our genetic toolkit. This work could advance selection for pigs that can better withstand disease or stress challenges. The outcome of similar heritability between stress environments is also encouraging, in that it suggests selection response is possible in both high stress and low stress environments. Planned future research will add historical phenotypes to strengthen our estimated trait relationships and help optimize the use of this information in the Genesus breeding program.
References:
Boyd D. (2012) Proc. of the Am. Assoc. of Swine Veterinarians, Perry City, IA United States.
Cheng J., Putz A.M., Harding J.C.S., Dyck M.K., Fortin F. et al. (2020) J Anim Sci. 98:8:1-14. https://doi.org/10.1093/jas/skaa244
Nguyen-Ba H., van Milgen J., Taghipoor M. (2020) Animal 14:2:253-260. https://doi.org/10.1017/S1751731119001976
Ocepek M., Andersen-Ranberg I., Edwards S. A., Fredriksen B., Framstad T. et al. (2016), J Anim Sci. 94:8:3550–3560. https://doi.org/10.2527/jas.2016-0386
Putz A.M., Harding J.C.S., Dyck M.K., Fortin F., Plastow G.S., Dekkers J.C.M. et al. (2019) Front. Genet. 9:660:1-14. https://doi.org/10.3389/fgene.2018.00660
Categorised in: Featured News, Global Tech
This post was written by Genesus



